Comparison of Compression Methods for Neuroimaging Data.
We are working on a pre-processing pipeline for a large neuroimaging dataset, and we want to be sure we are being judicious with our disk space usage. .nii Files are, conventionally, compressed with the program gzip (sometimes wrapped around a tape archive or tar file). Gzip is ubiquitously available, has a low memory footprint, and does an ok job. However, there are other perfectly mature, lossless compression formats available which get better results. If you are working with >100TB of data, this could matter a lot to your operating costs. Since compression performance is dependent on the type of data you had, I wanted to compare the efficiency of a number of algorithms and see what our options were.
Algorithms we are comparing.
Gzip and memcpy are included for comparison. Other compression tools were chosen based on their apparent popularity (from other compression tests published online or because of their inclusion in turbobench’s ‘standard lineups’) and to give a good range of datapoints from fast, minimally compressed to slow, highly compressed:
| method | level |
|---|---|
| brotli | 4 |
| brotli | 5 |
| bzip2 | N/A |
| flzma2 | 5 |
| flzma2 | 6 |
| flzma2 | 7 |
| flzma2 | 8 |
| flzma2 | 9 |
| libdeflate | 3 |
| libdeflate | 5 |
| libdeflate | 9 |
| lz4 | 1 |
| lzma | 5 |
| lzma | 6 |
| lzma | 7 |
| lzma | 8 |
| lzma | 9 |
| memcpy | N/A |
| zlib | 1 |
| zlib | 5 |
| zstd | 22 |
| zstd | 5 |
| zstd | 9 |
| gzip | N/A |
Blosc at level 11 was stopped manually after running for >12 hours.
Each tool was tested once on an HTPC instance with 1 processor and 8GB of memory. I additionally evaluated some methods with an instance with 4 processors and 32 GB of memory, but didn’t see large differences. Possibly Turbobench does not account for multithreading appropriately. I probably did not do this correctly – one thread is our target use case, so I did not spend a lot of time on multithreading.
Note that I am not positive the processors on the various HTPC servers used were identical, so there may be some noise in the timing data.
Tools
The tool I ended up using for most of the comparisons is called TurboBench, which has the advantages that it tests strictly in memory, has a lot of compression algorithms available, is flexible, and was easy for me to run on our HTPC cluster.
One thing Turbobench does not do is test gzip. Potentially one of the algorithms it offers is identical to gzip’s but I could not discern that, so I tested gzip using a separate script.
I was very curious about a library called blosc. Discussion on the github for NRRD suggested it might be ideal for this application. However, the lack of easily available command line tools for its use made me give up on it.
All these analyses were run at UW-Madison at CHTC using HTCondor. Code for analysis is available on the github repo.
Results
The full data table for this analysis is in the github repository as ‘fulldata.Rds’. I’m only going to plot points that are optimal on some dimension, and I’ll exclude a few outliers.
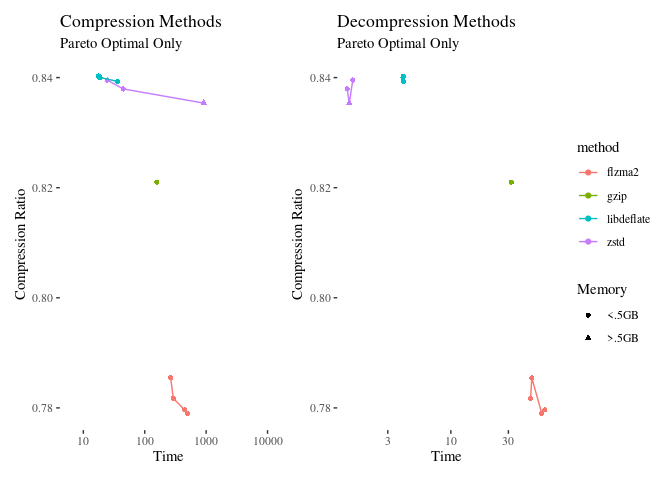
Discussion
In general, it is the compression benchmarks that seem to vary the most. Decompression is not much over 30 seconds even for the most time intensive method. flzma2 is a clear winner in these trials, with about 4% more compression than gzip. Flzma2 is not commonly available, and it would be best if we could use something less obscure. It is a fast implementation of LZMA, which is available in the package xz, so let’s compare those:
| method | clabel | ratio | ctime | |
|---|---|---|---|---|
| 4 | flzma2 | L5–37.0 MB | 0.8152 | 326 |
| 5 | flzma2 | L6–70.9 MB | 0.7855 | 263.8 |
| 6 | flzma2 | L7–138.8 MB | 0.7817 | 292.4 |
| 7 | flzma2 | L8–273.2 MB | 0.7796 | 446.8 |
| 8 | flzma2 | L9–273.2 MB | 0.779 | 492.4 |
| 13 | lzma | L5–168.3 MB | 0.797 | 438.9 |
| 14 | lzma | L6–336.0 MB | 0.7969 | 433.8 |
| 15 | lzma | L7–336.0 MB | 0.7969 | 672.3 |
| 16 | lzma | L8–604.5 MB | 0.795 | 689.7 |
| 17 | lzma | L9–604.5 MB | 0.795 | 906.2 |
Lzma at level 6 is within 1.5% of flzma2 at level 9, and is faster and uses less memory. So that’s probably our winner. It’s also the default setting of xz. As a bonus, xz supports integrity checking as a built in, which is very nice.
Here’s a plot of all the ‘lzma’ methods:
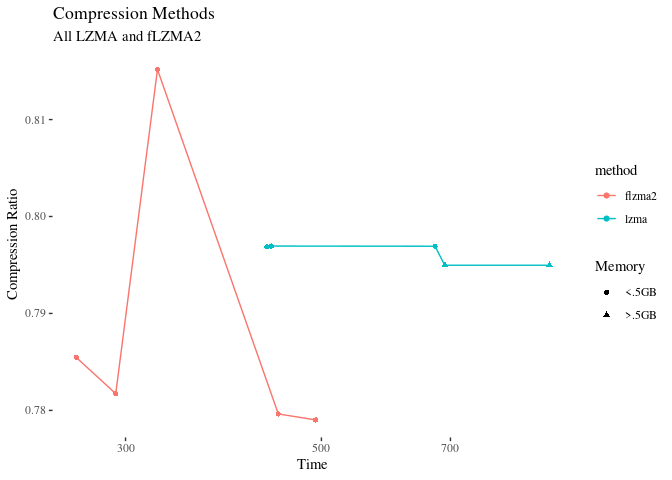
Mind the scales – the compression ratios are not actually that different here.
‘Real World’ testing
So the above testing is using just memory to memory compression, which is not the environment where our compression will actually happen. What about when we do this with disk i/o?
$ /usr/bin/time -f 'time: \t%e realSeconds \t%M peakMem' xz -zk subject.tar
time: 1525.79 realSeconds 97608 peakMem
$ ls -l subject.*
-rw-rw-r-- 1 peverill peverill 3045427200 Dec 16 09:37 subject.tar
-rw-rw-r-- 1 peverill peverill 2386532328 Dec 16 09:37 subject.tar.xz
So xz (lzma level 6) takes 25.4166667 minutes to compress the data, achieves a compression ratio of 0.7836445, and uses 97.6 MB of memory. It also appears to embed a file integrity check automatically. Sounds good!
What about Blosc?
The promise of Blosc for this type of data is that by using a pre-filter, it can better take advantage of the fact that a nifti file is ultimately an array of 16bit numbers, and the most significant digits don’t change that much (most compression algorithms do not account for this, but blosc’s pre-filtering options do). Don’t quote me on that, I’m following this forum post.
I tried a few times to get this working with various tools, but could not realize gains (certainly not to the extent to justify using a less mature tool).
With the compress_file program packaged with c-blosc2:
$ /usr/bin/time -f 'time: \t%e realSeconds \t%M peakMem' ./c-blosc2-2.6.0/build/examples/compress_file subject.tar subject.tar.b2frame
Blosc version info: 2.6.0 ($Date:: 2022-12-08 #$)
Compression ratio: 2904.3 MB -> 2710.9 MB (1.1x)
Compression time: 11.2 s, 260.3 MB/s
time: 11.15 realSeconds 5344 peakMem
With bloscpack using default options:
$ /usr/bin/time -f 'time: \t%e realSeconds \t%M peakMem' \
python3 packages/bin/blpk -v -n 1 c subject.tar
blpk: using 1 thread
blpk: getting ready for compression
blpk: input file is: 'subject.tar'
blpk: output file is: 'subject.tar.blp'
blpk: input file size: 2.84G (3045427200B)
blpk: nchunks: 2905
blpk: chunk_size: 1.0M (1048576B)
blpk: last_chunk_size: 354.0K (362496B)
blpk: output file size: 2.49G (2668748652B)
blpk: compression ratio: 1.141144
blpk: done
time: 8.15 realSeconds 44392 peakMem
The same, but using the zstd algorithm:
$ /usr/bin/time -f 'time: \t%e realSeconds \t%M peakMem' python3 packages/bin/blpk -vn 1 c --codec zstd subject.tar
blpk: using 1 thread
blpk: getting ready for compression
blpk: input file is: 'subject.tar'
blpk: output file is: 'subject.tar.blp'
blpk: input file size: 2.84G (3045427200B)
blpk: nchunks: 2905
blpk: chunk_size: 1.0M (1048576B)
blpk: last_chunk_size: 354.0K (362496B)
blpk: output file size: 2.15G (2306001080B)
blpk: compression ratio: 1.320653
blpk: done
time: 134.08 realSeconds 51328 peakMem
Finally, to make sure that I was using bit-shuffling (which is supposedly where the magic happens), I wrote a custom version of the compress_file program. Assuming I did that right, here is the output:
$ /usr/bin/time -f 'time: \t%e realSeconds \t%M peakMem' c-blosc2-2.6.0/build/examples/compress_file subject.tar subject.tar.b2frame
Blosc version info: 2.6.0 ($Date:: 2022-12-08 #$)
Compression ratio: 2904.3 MB -> 2397.1 MB (1.2x)
Compression time: 52.3 s, 55.5 MB/s
time: 52.34 realSeconds 9084 peakMem
In fairness, the best version (zstd using bloscpack) compressed the file at 75.7% in just over two minutes, using 51MB of ram – much superior to lzma. Also, all of these tests used typesize=8, and possibly it should be 16. However, it’s not enough of a benefit to justify the additional complexity (and I ran out of time exploring it).