On transporting FSL designs to SPM
Introduction
Let’s say, just hypothetically, that you built a first-level analysis pipeline in FSL but are then find you need to run SPM code on it. You might wish there was a way to easily convert your design in to an SPM readable format. Here are a few notes on that process, along with a python script to actually output a timing.mat file:
Inputting 4d data
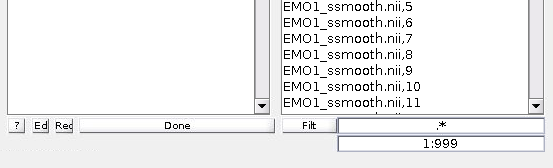
If you are used to working with FSL, you are probably using 4d .nii files. Fortunately, SPM8 can open these (my understanding is that this didn’t used to be the case). BUT you have to specify which volumes of the files you want to examine. So if your scan selection says ‘Run1.nii,1’ - that means you’ve only selected the first volume. Your batch is going to fail with ‘Cell contents reference from a non-cell array object.’ which might, hypothetically, cause you to spend a few days wondering what is wrong with your timing file. That would be frustrating.
Instead, what you need to do is change the window that reads ‘1’ under the filter box in the file selection window to 1:999, like so:
You will see all the volumes appear in the list. Select them all and press ‘Done’. Now you are ready to go!
The SPM Wikibook has more info on working with 4d data
Adding your timings
So, using the GUI, you can enter your conditions with names, onsets, and durations as separate vectors, but that’s annoying. You can also specify a .mat file containing three cell arrays with those values, which is less annoying but you still have to transcribe from your carefully produced three column FSL event file. If only you had a script to do that for you…
Here is that script:
#This script takes any number of three column fsl event files and outputs a .m matlab script suitable for generating timing parameters.
import sys
import pandas
#from decimal import Decimal
print 'Number of arguments:', len(sys.argv[1:]), 'arguments.'
print 'Argument List:', str(sys.argv[1:])
llist=['names=cell(1,%s);' % len(sys.argv[1:]),
'onsets=cell(1,%s);' % len(sys.argv[1:]),
'durations=cell(1,%s);' % len(sys.argv[1:])]
#Get the EVs from the fsl files.
for i in enumerate(sys.argv[1:], start=1):
try:
f = open(i[1])
except:
print "Could not open file %s" % i[1]
exit(1)
df = pandas.read_csv(f, delim_whitespace=True, header=None).sort_index(by=0)
llist.append("names{%s}='%s';" % i)
#This is awful, sorry:
llist.append("onsets{%s}=[%s];" % (i[0], ', '.join(["{0:.4f}".format(o) for o in df[0].tolist()])))
llist.append("durations{%s}=%s;" % (i[0], str(df[1].tolist())))
f.close()
w = open('output.m','w')
print 'output.m file text'
for l in llist:
print l
w.write("%s\n" % l)
w.close()
print "\nWrote output.m"
#print(str(llist)) To use this, just run this from the command line:
python fslevsToMat.py fslevfile1.txt fslevfile2.txtIt will create a file ‘output.m’ in the working directory. You need to run this script within matlab and then save the resulting cell matrices as a .mat file, then input that file in matlab.
If you need to run the script a lot, consider adding a #! line specifying your local python executable and placing it somewhere within your path variable, so you can just run it.]]>